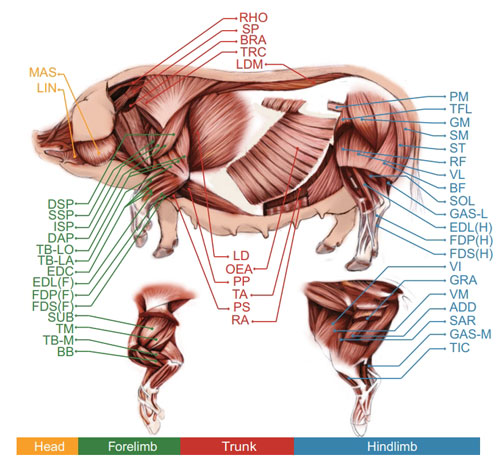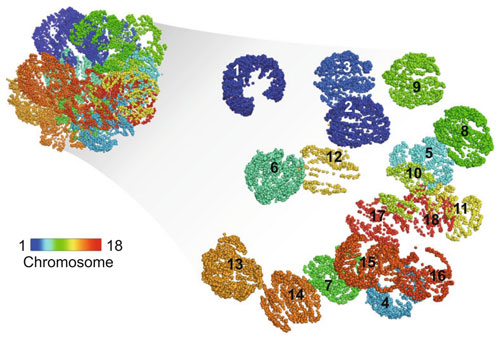
The dynamic interactions between noncoding regulatory elements and polygenes shaped by the three-dimensional (3D) organization of chromosomes provide adequate explanations for the molecular basis of muscle growth. Our group closely aligned with 'multi-omics and big data analysis for the development of efficient and precise breeding technology' , with a focus on the potential effects of genomic variations on the disruption of chromatin organization thus regulating gene transcription.
We will further provide the functional and biological evidences for the candidate variations in porcine primary cell and based on the mouse model, as well as the gene edited pigs, and assess their potential applications in molecular breeding. We envision that our discoveries will provide new information from a non-coding region perspective to reveal the complex molecular mechanisms of muscle growth traits, providing a theoretical basis for the development of efficient pig breeding technology and precision improvement.